The Advantages of NeXGen Fungal / AFB NGS Testing
Eurofins Viracor has developed and validated a first-of-its-kind next generation sequencing assay. NeXGen™ hybrid-capture NGS emerges as a superior choice in infectious disease identification. Hybrid-capture NGS is a powerful technique that combines the specificity of target capture with the high-throughput capabilities of NGS offering advantages such as:
- Comprehensive Pathogen Detection: Traditional diagnostic methods often rely on culture-based techniques, which can be time-consuming and limited in their ability to detect fastidious or slow-growing organisms. Hybrid-capture NGS overcomes these limitations by providing a comprehensive view of the microbial community present in a specimen, enabling the detection of both known and novel pathogens.
- Increased Sensitivity and Specificity: The use of specific probes in hybrid-capture NGS enhances the sensitivity and specificity of pathogen detection. By targeting conserved regions of the pathogen's genome, this technique can accurately identify even low-abundance organisms, reducing the risk of false-negative results.
- Simultaneous Detection of Multiple Pathogens: Hybrid-capture NGS allows for the simultaneous detection of multiple pathogens in a single assay. This is particularly advantageous in cases where co-infections or mixed infections are suspected, as it provides a comprehensive overview of the pathogens present, aiding in accurate diagnosis and appropriate treatment selection.

NeXGen™ Fungal / AFB NGS Testing
NeXGen hybrid-capture NGS offers a powerful and comprehensive approach to infectious disease testing, providing clinicians with valuable insights on prominent or “low load” clinically relevant microorganisms allowing for a more accurate diagnosis and appropriate treatment decisions.
NeXGen™ Fungal / AFB NGS (WB/Plasma) NeXGen™ Fungal / AFB NGS (BAL)
hybrid-capture next generation sequencing
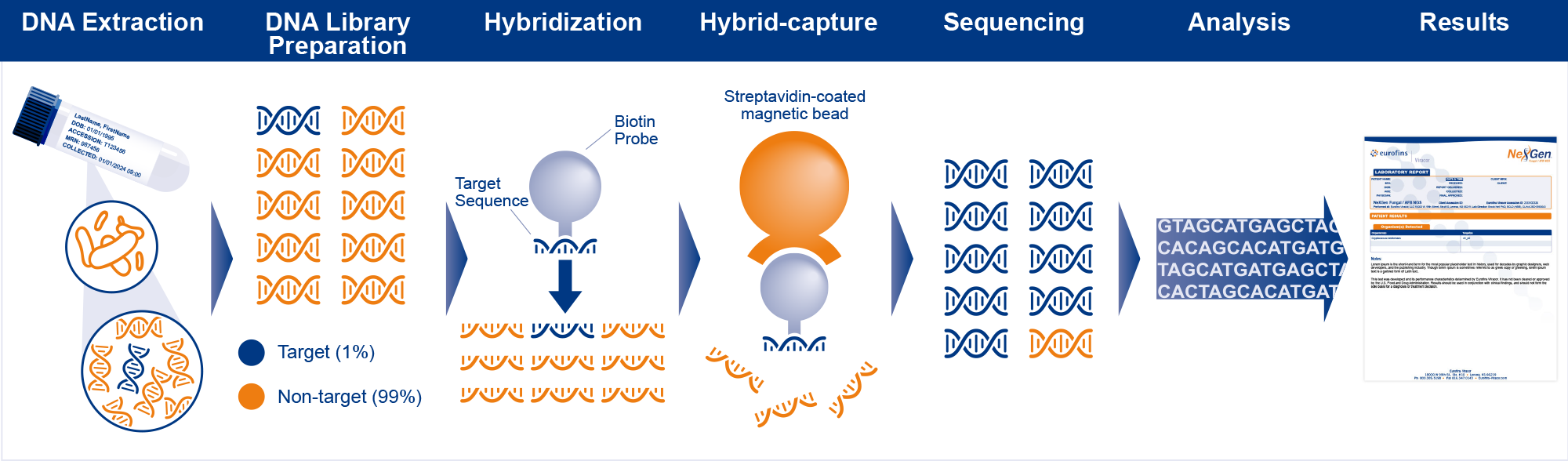
How It Works
Organisms Dataset
Click for a detailed Organism Dataset PDF list
Absidia (26)
Actinomucor (1)
Actinomyces (40)
Aggregatibacter (1)
Alternaria (3)
Apiotrichum (1)
Apophysomyces (3)
Aspergillus (42)
Aureobasidium (4)
Basidiobolus (2)
Blastomyces (3)
Candida (25)
Chaetomium (1)
Chrysosporium (1)
Cladophialophora (5)
Cladosporium (1)
Clavispora (1)
Coccidioides (2)
Cokeromyces (1)
Conidiobolus (1)
Coniosporium (1)
Cryptococcus (2)
Cryptococcus gattii VGI (1)
Cryptococcus gattii VGII (1)
Cryptococcus gattii VGIII (1)
Cunninghamella (11)
Curvularia (2)
Cutaneotrichosporon (1)
Cyberlindnera (2)
Cyphellophora (1)
Debaryomyces (2)
Diutina (1)
Emergomyces (5)
Emmonsia (1)
Enterocytozoon (1)
Exophiala (7)
Filobasidium (1)
Fonsecaea (5)
Fusarium (25)
Geotrichum (1)
Histoplasma (1)
Hyphopichia (1)
Kluyveromyces (2)
Leptosphaeria (1)
Lichtheimia (2)
Lodderomyces (1)
Lomentospora (1)
Madurella (1)
Malassezia (13)
Memnoniella (1)
Metarhizium (7)
Metschnikowia (2)
Meyerozyma (2)
Microsporum (1)
Mortierella (1)
Mucor (5)
Mycobacterium (115)
Mycobacterium abscessus (4)
Mycobacterium avium (1)
Mycobacterium avium subsp. paratuberculosis (1)
Mycobacterium avium subsp. silvaticum (1)
Mycobacterium intracellulare (1)
Mycobacterium intracellulare subsp. chimaera (1)
Mycobacterium intracellulare subsp. (1) yongonense
Mycobacterium tuberculosis Complex (5)
Mycobacteroides abscessus (1)
Mycobacteroides abscessus
subsp. bolletii (1)
Mycobacteroides abscessus subsp. (1) massiliense
Mycobacteroides (1)
Mycolicibacter (2)
Mycolicibacterium (18)
Mycolicibacterium fortuitum (3)
Nakaseomyces (2)
Nannizzia (1)
Nigrograna (1)
Nocardia (98)
Nocardiopsis (1)
Paecilomyces (1)
Paracoccidioides (2)
Pascua (1)
Penicillium (23)
Phaeoacremonium (1)
Phaeotremella (2)
Phialophora (1)
Pichia (1)
Pneumocystis (3)
Podila (1)
Purpureocillium (1)
Rasamsonia (13)
Rhinocladiella (1)
Rhizomucor (2)
Rhizopus (5)
Rhodotorula (3)
Rhytidhysteron (1)
Saccharomyces (1)
Saksenaea (2)
Samsoniella (1)
Scedosporium (4)
Schaalia (3)
Schizophyllum (1)
Sporothrix (4)
Stachybotrys (2)
Starmerella (2)
Suhomyces (1)
Syncephalastrum (2)
Talaromyces (11)
Thermoascus (1)
Thermothelomyces (1)
Trichoderma (9)
Trichophyton (6)
Trichosporon (2)
Ustilago (5)
Verruconis (7)
Wallemia (2)
Wickerhamiella (2)
Wickerhamomyces (2)
Yamadazyma (1)
Yarrowia (1)
B R O C H U R E
The Eurofins Viracor NeXGen™ Hybrid-capture Fungal/AFB NGS Assay


Hybrid-capture NGS is a powerful technique that combines the specificity of target capture with the high-throughput capabilities of NGS
It offers advantages such as:
- Comprehensive Pathogen Detection
- Increased Sensitivity and Specificity
- Simultaneous Detection of Multiple Pathogens
Read More...
Hybrid-Capture NGS for Detecting Fungal and Acid-Fast Bacteria
NeXGen™ Fungal / AFB NGS Assay
Next Generation Sequencing (NGS) testing has gained significant importance in the medical clinical laboratory in the United States over the past decade. In recent years, the field of molecular diagnostics has witnessed significant advancements, particularly in the detection of infectious diseases. One such breakthrough is the application of hybrid-capture next-generation sequencing (NGS) technology.
- Hybrid-capture NGS testing combines the targeted approach of 16s DNA sequencing with the comprehensive nature of NGS, enabling the analysis of specific genomic regions of interest.
33356 - NeXGen™ Fungal / AFB NGS (WB/Plasma) Assay NeXGen™ Fungal / AFB NGS (BAL)

- Singh, Rajesh R. “Target Enrichment Approaches for Next-Generation Sequencing Applications in Oncology.” Diagnostics (Basel, Switzerland) vol. 12,7 1539. 24 Jun. 2022, doi:10.3390/diagnostics12071539
